Introduction
Welcome to Genestrip Web!
Genestrip Web is a software for metagenomic analysis of DNA sequencing data.
Through a simple web interface, you can perform the corresponding
analysis without the need for training or specialized IT knowledge.
Genestrip Web consists of three components:
- The web-based user interface with a subsystem for processing DNA data.
- The actual analysis program called GeneStrip.
- A collection of so-called databases to assign DNA fragments to the appropriate organisms.
Genestrip Web requires input files for analysis in fastq format — as so-called fastq files.
These must first be generated with a DNA sequencer from an environmental or tissue sample.
This step is complex and is usually carried out by trained personnel in biological or genetic laboratories.
In contrast, the subsequent analysis of fastq files with GeneStrip Web is straightforward.
The result of an analysis with GeneStrip Web is a CSV file, which you can open, for example, with MS Excel or view directly within GeneStrip Web.
The file contains various statistical values indicating which organisms are detected in the fastq file.
From this, you can often draw conclusions about the organisms present in the sample.
The following diagram illustrates the entire process schematically.
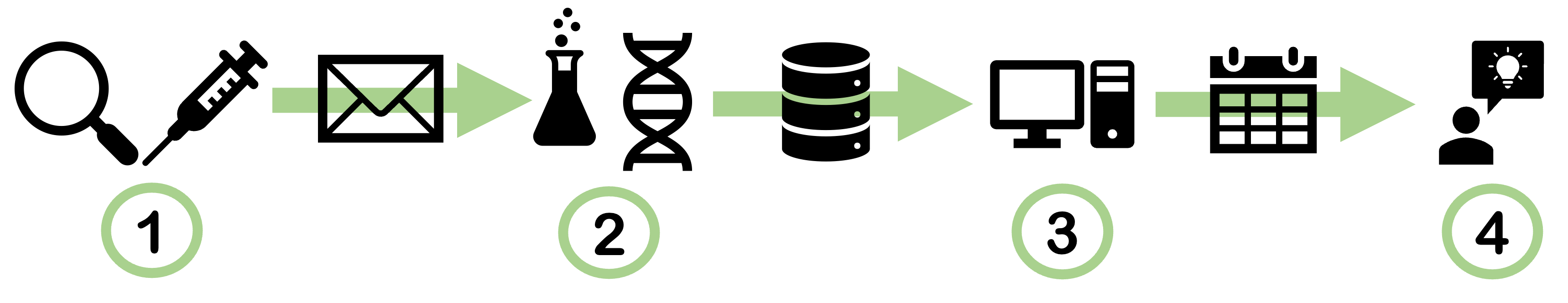
- The sampling provides an environmental or tissue sample.
- DNA sequencing of the sample yields one or more fastq files.
- The analysis with GeneStrip Web generates CSV files.
- The CSV files are used by researchers and experts to draw conclusions about the organisms present in the sample.
Important: An analysis using Genestrip Web or GeneStrip is not suitable for medical diagnostics!
Online Demo
You can find an online demo of Genestrip Web here:
https://genestrip.it.hs-heilbronn.de/gweb
The online demo shows, among other things, analysis results for the examination of 6 ticks based on the Nature publication “Metagenomic surveillance for bacterial tick-borne pathogens using nanopore adaptive sampling”.
For this, the fastq files from the publication were analyzed with Genestrip Web. All corresponding results from the publication were reproducible.
You can view them without logging in in the online demo. You can also view information about some of the available databases without logging in.
Analyses cannot be performed in the online demo without registration.
Benefits at a Glance
- Genestrip Web can be installed and run with just a few clicks on common, modern Windows PCs, while other metagenomic analysis tools mainly operate on Unix-based systems. (Genestrip Web can also run on Macs, Linux, etc.)
- Genestrip Web provides an easy-to-use web interface, whereas other metagenomic tools require command-line operations.
(However, the core system GeneStrip can still be used from the command line.) - Genestrip Web offers an interesting and relevant collection of databases for easy installation, covering many human pathogens.
Additional databases can be generated by IT administrators with moderate effort using GeneStrip. - Compared to alternative systems, such as KrakenUniq, GeneStrip Web requires relatively little main memory for specialized analysis databases.
- In contrast to, for example, Kraken2, GeneStrip Web provides very accurate and comprehensive analysis results for assigned organisms.
License
Genestrip Web or for short “gweb” is free for non-commercial use under this license.
 Heilbronn University
Heilbronn University